That can be easily eliminatedAn artistic decision tree generated by Dall-EThere is a chance that your decision trees and random forests suffer from a small bias, which can be eliminated with ease, for basically no cost. This is what we...
That can be easily eliminated
 An artistic decision tree generated by Dall-E
An artistic decision tree generated by Dall-EThere is a chance that your decision trees and random forests suffer from a small bias, which can be eliminated with ease, for basically no cost. This is what we explore in this post.
Disclaimer: the post discusses a recent research conducted by the author.
Introduction
Decision trees and random forests are widely adopted classification and regression techniques in machine learning. Decision trees are favored for their interpretability, while random forests stand out as highly competitive and general-purpose state-of-the-art techniques. Commonly used CART implementations, such as those in the Python package sklearn and the R packages tree and caret, assume that all features are continuous. Despite this silent assumption of continuity, both techniques are routinely applied to datasets with diverse feature types.
In a recent paper, we investigated the practical implications of violating the assumption of continuity and found that it leads to a bias. Importantly, these assumptions are almost always violated in practice. In this article, we present and discuss our findings, illustrate and explain the background, and propose some simple techniques to mitigate the bias.
A motivating example
Let’s jump into it with an example using the CPU performance dataset from the UCI repository. We’ll import it through the common-datasets package, to simplify the preprocessing and bypass the need for feature encoding and missing data imputation.
import numpy as npfrom sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import RepeatedKFold, cross_validate
from scipy.stats import wilcoxon
from common_datasets.regression import load_cpu_performance
dataset = load_cpu_performance()
X = dataset['data']
y = dataset['target']
# a cross-validation wrapper to simplify the code
def cv_rf(X, y, regressor=RandomForestRegressor):
return cross_validate(
estimator=regressor(max_depth=11),
X=X, y=y,
cv=RepeatedKFold(n_splits=5, n_repeats=400, random_state=5),
scoring='r2'
)['test_score']
r2_original = cv_rf(X, y)
r2_mirrored = cv_rf(-X, y)
In the experiment, we evaluate the performance of the random forest regressor on both the original data and its mirrored version (each feature multiplied by -1). The hyperparameter for the regressor (max_depth=11 ) was chosen in a dedicated model selection step, maximizing the r2 score across a reasonable range of depths. The cross-validation employed for evaluation is significantly more comprehensive than what is typically used in machine learning, encompassing a total of 2000 folds.
print(f'original r2: {np.mean(r2_original):.4f}')print(f'mirrored r2: {np.mean(r2_mirrored):.4f}')
print(f'p-value: {wilcoxon(r2_original, r2_mirrored, zero_method="zsplit").pvalue:.4e}')
# original r2: 0.8611
# mirrored r2: 0.8595
# p-value: 6.2667e-04
In terms of r2 scores, we observe a deterioration of 0.2 percentage points when the attributes are mirrored. Furthermore, the difference is statistically significant at conventional levels (p << 0.01).
The results are somewhat surprising and counter-intuitive. Machine learning techniques are typically invariant to certain types of transformations. For example, k Nearest Neighbors is invariant to any orthogonal transformation (like rotation), and linear-ish techniques are typically invariant to the scaling of attributes. Since the space partitioning in decision trees is axis aligned, it cannot be expected to be invariant to rotations. However, it is invariant to scaling: applying any positive multiplier to any feature will lead to the exact same tree. Consequently, there must be something going on with the mirroring of the axes.
An intriguing question arises: what if mirroring the axes leads to better results? Should we consider another degree of freedom (multiplying by -1) in model selection beyond determining the optimal depth? Well, in the rest of the post we figure out what is going on here!
Binary decision tree induction and inference
Now, let’s briefly review some important characteristics of building and making inferences with binary Classification And Regression Trees (CART), which are used by most implementations. A notable difference compared to other tree induction techniques like ID3 and C4.5 is that CART trees do not treat categorical features in any special way. CART trees assume that all features are continuous.
Given a training set (classification or regression), decision trees are induced by recursively partitioning the training set alongside the feature space using conditions like feature < threshold or alternatively, features <= threshold. The choice of conditioning is usually an intrinsic property of the implementations. For example, the Python package sklearn uses conditions of the form feature <= threshold, while the R package tree uses feature < threshold. Note that these conditions are aligned with the presumption of all features being continuous.
Nevertheless, the presumption of continuous features is not a limitation. Integer features, category features through some encoding, or binary features can still be fed into these trees. Let’s examine an example tree in a hypothetical loan approval scenario (a binary classification problem), based on three attributes:
graduated (binary): 0 if the applicant did not graduate, 1 if the applicant graduated;income (float): the yearly gross income of the applicant;dependents (int): the number of dependents;and the target variable is binary: whether the applicant defaults (1) or pays back (0).
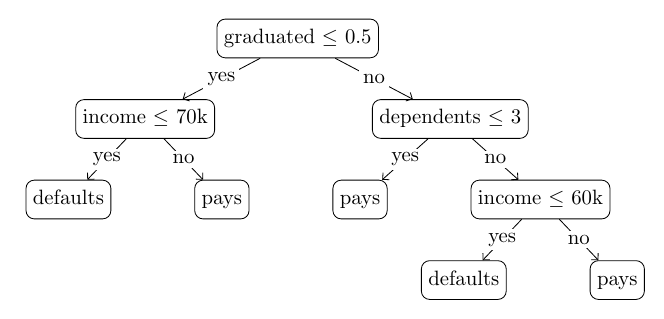 A decision tree built for a hypothetical loan approval scenario
A decision tree built for a hypothetical loan approval scenarioThe structure of the tree, as well as the conditions in the nodes (which threshold on which feature), are inferred from the training data. For more details about tree induction, refer to decision tree learning on Wikipedia.
Given a tree like this, inference for a new record is conducted by starting from the leaf node, recursively applying the conditions, and routing the record to the branch corresponding to the output of the condition. When a leaf node is encountered, the label (or eventually distribution) recorded in the leaf node is returned as the prediction.
The conditioning and the threshold
A finite set of training records cannot imply a unique partitioning of the feature space. For example, the tree in the figure above could be induced from data where there is no record with graduation = 0 and income in the range ]60k, 80k[. The tree induction method identifies that a split should be made between the income values 60k and 80k. In the absence of further information, the midpoint of the interval (70k) is used as the threshold. Generally, it could be 65k or 85k as well. Using the midpoints of the unlabeled intervals is a common practice and a reasonable choice: in line with the assumption of continuous features, 50% of the unlabeled interval is assigned to the left and 50% to the right branches.
Due to the use of midpoints as thresholds, the tree induction is completely independent of the choice of the conditioning operator: using both <= and < leads to the same tree structure, with the same thresholds, except for the conditioning operator.
However, inference does depend on the conditioning operator. In the example, if a record representing an applicant with a 70k income is to be inferred, then in the depicted setup, it will be routed to the left branch. However, using the operator <, it will be routed to the right branch. With truly continuous features, there is a negligible chance for a record with exactly 70k income to be inferred. However, in reality, the income might be quoted in units of 1k, 5k, or eventually 10k, which makes it probable that the choice of the conditioning operator has a notable impact on predictions.
The relation of conditioning and mirroring
Why do we talk about the conditioning when the problem we observed is about the mirroring of features? The two are basically the same. A condition “feature < threshold” is equivalent to the condition “-feature <= -threshold” in the sense that they lead to the same, but mirrored partitioning of the real axis. Namely, in both cases, if the feature value equals the threshold, that value is in the same partition where the feature values greater than the threshold are. For example, compare the two trees below, the one we used for illustration earlier, except all conditioning operators are changed to <, and another tree where the operator is kept, but the tree is mirrored: one can readily see that for any record they lead to the same predictions.
 The previous tree with the conditioning operator <
The previous tree with the conditioning operator <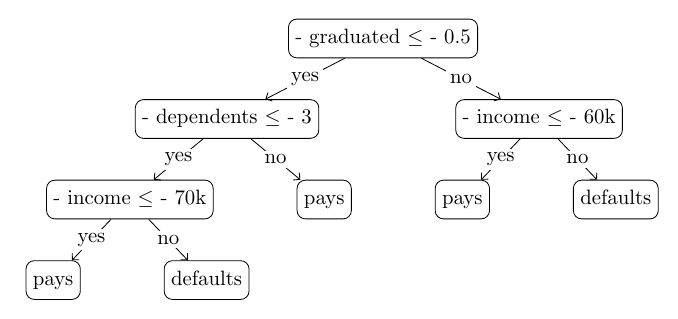 The tree built on the mirrored data (still using the conditioning operator ?)
The tree built on the mirrored data (still using the conditioning operator ?)Since tree induction is independent of the choice of conditioning, building a tree on mirrored data and then predicting mirrored vectors is equivalent to using the non-default conditioning operator (<) for inference on non-mirrored records. When the trees of the forest were fitted to the mirrored data, even though sklearn uses the ‘<=’ operator for conditioning, it worked as if it used the ‘<’ operator. Consequently, the performance deterioration we discovered with mirroring is due to thresholds coinciding with feature values, leading to different predictions during the evaluation of the test sets.
For the sake of completeness, we note that the randomization in certain steps of tree induction might lead to slightly different trees when fitted to the mirrored data. However, these differences smooth out in random forests, especially in 2k folds of evaluation. The observed performance deterioration is a consequence of the systematic effect of thresholds coinciding with feature values.
When can this happen?
Primarily, two circumstances increase the likelihood of the phenomenon:
When a feature domain contains highly probable equidistant values: This sets the stage for a threshold (being the mid-point of two observations) to coincide with a feature value with high probability.Relatively deep trees are built: Generally, as a tree gets deeper, the training data becomes sparser at the nodes. When certain observations are absent at greater depths, thresholds might fall on those values.Interestingly, features taking a handful of equidistant values are very common in numerous domains. For example:
The age feature in medical datasets.Rounded decimals (observed to the, say, 2nd digit will form a lattice).Monetary figures quoted in units of millions or billions in financial datasets.Additionally, almost 97% of features in the toy regression and classification datasets in sklearn.datasets are of this kind. Therefore, it is not an over-exaggeration to say that features taking equidistant values with high probability are present everywhere. Consequently, as a rule of thumb, the deeper trees or forests one builds, the more likely it becomes that thresholds interfere with feature values.
It is a bias, model selection cannot help!
We have seen that the two conditioning operators (the non-default one imitated by the mirroring of data) can lead to different prediction results with statistical significance. The two predictions cannot be unbiased at the same time. Therefore, we consider the use of either form of conditioning introducing a bias when thresholds coincide with feature values.
Alternatively, it is tempting to consider one form of conditioning to be luckily aligned with the data and improving the performance. Thus, model selection could be used to select the most suitable form of conditioning (or whether the data should be mirrored). However, in a particular model selection scenario, using some k-fold cross-validation scheme, we can only test which operator is typically favorable if, say, 20% of the data is removed (5-fold) from training and then used for evaluation. When a model is trained on all data, other thresholds might interfere with feature values, and we have no information on which conditioning would improve the performance.
Mitigating the bias in random forests
A natural way to eliminate the bias is to integrate out the effect of the choice of conditioning operators. This involves carrying out predictions with both operators and averaging the results.
In practice, with random forests, exploiting the equivalence of data mirroring and changing the conditioning operator, this can be approximated for basically no cost. Instead of using a forest of N_e estimators, one can build two forests of half the size, fit one to the original data, the other to the mirrored data, and take the average of the results. Note that this approach is applicable with any random forest implementation, and has only marginal additional cost (like multiplying the data by -1 and averaging the results).
For example, we implement this strategy in Python below, aiming to integrate out the bias from the sklearn random forest.
from sklearn.base import RegressorMixinclass UnbiasedRandomForestRegressor(RegressorMixin):
def __init__(self, **kwargs):
# determining the number of estimators used in the
# two subforests (with the same overall number of trees)
self.n_estimators = kwargs.get('n_estimators', 100)
n_leq = int(self.n_estimators / 2)
n_l = self.n_estimators - n_estimators_leq
# instantiating the subforests
self.rf_leq = RandomForestRegressor(**(kwargs | {'n_estimators': n_leq}))
self.rf_l = RandomForestRegressor(**(kwargs | {'n_estimators': n_l}))
def fit(self, X, y, sample_weight=None):
# fitting both subforests
self.rf_leq.fit(X, y, sample_weight)
self.rf_l.fit(-X, y, sample_weight)
return self
def predict(self, X):
# taking the average of the predictions
return np.mean([self.rf_leq.predict(X), self.rf_l.predict(-X)], axis=0)
def get_params(self, deep=False):
# returning the parameters
return self.rf_leq.get_params(deep) | {'n_estimators': self.n_estimators}
Next, we can execute the same experiments as before, using the exact same folds:
r2_unbiased = cv_rf(X, y, UnbiasedRandomForestRegressor)Let’s compare the results!
print(f'original r2: {np.mean(r2_original):.4f}')print(f'mirrored r2: {np.mean(r2_mirrored):.4f}')
print(f'unbiased r2: {np.mean(r2_unbiased):.4f}')
# original r2: 0.8611
# mirrored r2: 0.8595
# unbiased r2: 0.8608
According to expectations, the r2 score of the unbiased forest falls between the scores achieved by the original forest with and without mirroring the data. It might seem that eliminating the bias is detrimental to the performance; however, we emphasize again that once the forest is fit with all data, the relations might be reversed, and the original model might lead to worse predictions than the mirrored model. Eliminating the bias by integrating out the dependence on the conditioning operator eliminates the risk of deteriorated performance due to relying on the default conditioning operator.
Conclusion
The presence of a bias related to the interaction of the choice of conditioning and features taking equidistant values has been established and demonstrated. Given the common occurrence of features of this kind, the bias is likely to be present in sufficiently deep decision trees and random forests. The potentially detrimental effect can be eliminated by averaging the predictions carried out by the two conditioning operators. Interestingly, in the case of random forests, this can be done at basically no cost. In the example we used, the improvement reached the level of 0.1–0.2 percentage points of r2 scores. Finally, we emphasize that the results generalize to classification problems and single decision trees as well (see preprint).
Further reading
For further details, I recommend:
Our preprint discussing the topic in detail, with many more tests, illustrations and alternative bias mitigation techniques: preprint,The GitHub repository with the reproducible analysis: https://github.com/gykovacs/conditioning_biasThe notebook behind this post: https://github.com/gykovacs/conditioning_bias/blob/main/blogpost/001-analysis.ipynbIf you are interested in more content like this, don’t forget to subscribe! You can also find me on LinkedIn!
A Subtle Bias that Could Impact Your Decision Trees and Random Forests was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.